Using the Mutated Protein to Verify Our COVID-19 Antibodies Pairs
The antibodies pairs (ZLA81120M +ZLA81122H) can recognize and react well with B.1.1.7, B.1.351, B.1.2, P1, P2, B.1.617.2, B.1.617.3, and C.37.
1)The full-length structure of the COVID-19 N protein:
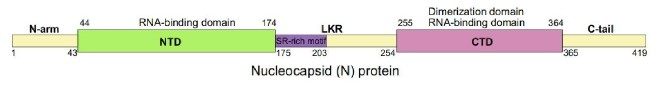
According to the full-length structure of the COVID-19 N protein, the full-length Protein is divided into fragments of different regions for expression, including N-Arm, NTD, LKR-SR, LKR-R, CTD, C-Tai, as well as N-Arm- NTD, NTD-LKR-SR, LKR, LKR-R-CTD, CTD-C-Tail. And the scientists preliminarily analyzed the approximate area of the antibody epitope.
The epitope of the coated antibody is 44-54, but the structure of the LKR-SR position will affect the binding of the antibody to the antigen. The labelled antibody epitope is a conformational epitope that is located at NTD. The core region is 149-178, which the amino acid affects at positions 104-149. The experiment successfully verifies that the antibody pairs can detect the mutations in the NTD position by analyzing the presently popular mutant strains. In contrast, the other mutations are not in the NTD position.
2) The result of detection of variant strains:
Strains | Repeat 1 | Repeat 2 | N protein mutation site |
B.1.1.7 (Alpha) | + | + | D3L, R203K, G204R, S235F |
B.1.351 (Beta) | + | + | T205I |
B.1.2 | + | + | P67S, P199L |
P.1(Gamma) | + | + | P80R |
P.2 | + | + | A119S, M234I |
B.1.617.2 (Delta) | + | + | R203M, D377Y |
B.1.617.3 | + | + | P67S, R203M, D377Y |
C.37 (Lambda) | + | + | P13L, R203K, G204R, G214C |
3) Result Analysis
The antibodies pairs (ZLA81120M +ZLA81122H)can recognize and react well with B.1.1.7, B.1.351, B.1.2, P1, P2, B.1.617.2, B.1.617.3, and C.37.
Product Name | Cat.NO. | Source | Clone | Product Description |
Mouse Anti-2019-ncov NP Antibody | ZLA81120M | Mouse IgG | 3F6M | LFA,CLIA coating |
Humanized Anti-2019-ncov NP antibody | ZLA81122H | Human IgG | 7G2H | LFA,CLIA, ELISA labeling |
4) Quality Control Material
Cat.NO. | Product Name | Product Description |
ZLP8118K | Recombinant nCoV NP antigen control,D3L,S235F mutation | HEK293,no tag,B.1.17 variant |
ZLP8119K | Recombinant nCoV NP antigen control,R203K,G204R | HEK293,no tag,B.1.519 |
ZLP81110K | Recombinant nCoV NP antigen control,P67S,P199L mutation | HEK293,no tag,B.1.2 variant |
ZLP81112K | Recombinant nCoV NP antigen control,RA119S,M234I mutation | HEK293,no tag,P.2 variant |
ZLP81116K | Recombinant nCoV NP antigen control,D3L, R203K,G204R,S235F mutation | HEK293,no tag,B.1.17 variant(Alpha) |
ZLP81111K | Recombinant nCoV NP antigen control,P80R mutation | HEK293,no tag,P.1variant(Gamma) |
ZLP81117K | Recombinant nCoV NP antigen control,T205I mutation | HEK293,no tag,B.1.351variant(Beta) |
ZLP81113K | Recombinant nCoV NP antigen control,R203M,D377Y mutation | HEK293,no tag,B.1.617.2 variant(Delta) |
ZLP81114K | Recombinant nCoV NP antigen control,P67S,R203M,D377Y mutation | HEK293,no tag,B.1.167.3variant |
ZLP81115K | Recombinant nCoV NP antigen control,P13L,R203K,G204R,G214Cmutation | HEK293,no tag,C37 variant (Lambda) |
Related Immunoassays
- Cardiac Markers
-
Tumor Marker
-
PGII
-
G17
- CA50
-
CA125
- CA242
-
CA15-3
- CA19-9
- CA72-4
-
Pepsinogens I (PGI)
-
Human Epididymis 4 (HE4)
- Prostate-Specific Antigen (PSA)
- Squamous Cell Carcinoma (SCC)
- Neuron-Specific Enolase (NSE)
- Cytokeratin 19 Fragment (CYFRA21-1)
- Human Progastrin-releasing Peptide (ProGRP Tumor Marker)
- Protein Induced by Vitamin K Absence or Antagonist-II (PIVKA II Tumor Marker)
- Alpha-fetoprotein(AFP)
-
CEA
-
Human Chitinase 3-like 1
-
PGII
- Inflammatory Marker
- Infectious Disease
- Hormones
- Thyroid Function
- Glucose Metabolism
- Bone Marker
- Others
-
Heterophilic Blocking Reagent
- Animal Diagnostics