Myxovirus-Resistant Protein A (MxA)
Mx gene was found in 1964 and was named after its resistance to myxovirus. After the viral infection, the Mx gene is induced by type I interferon-IFN (α/β) to produce MxA and MxB proteins. Both are distributed in the cytoplasm. Mx proteins are members of the family of large GTPases and central innate immune system players.
MxA proteins mediate cellular resistance against various pathogens, including influenza viruses and many other related viruses. MxA proteins can bind to the viral capsid and inhibit viral transcription and translation. MxA proteins are closely associated with the viral infection and can be used for early diagnosis of viral infection.
Myxovirus-resistant protein A (MxA Antibody) Products
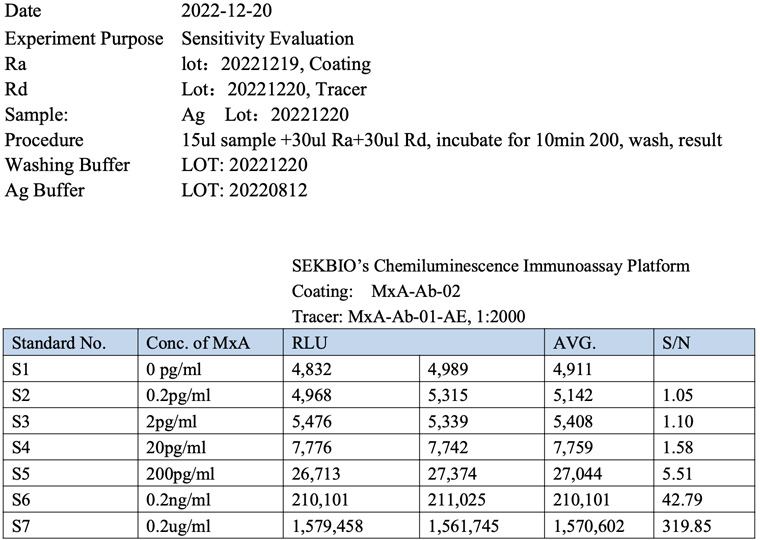
The specificity of MxA Master sheet, feedback from our customer
Structure of MxA proteins
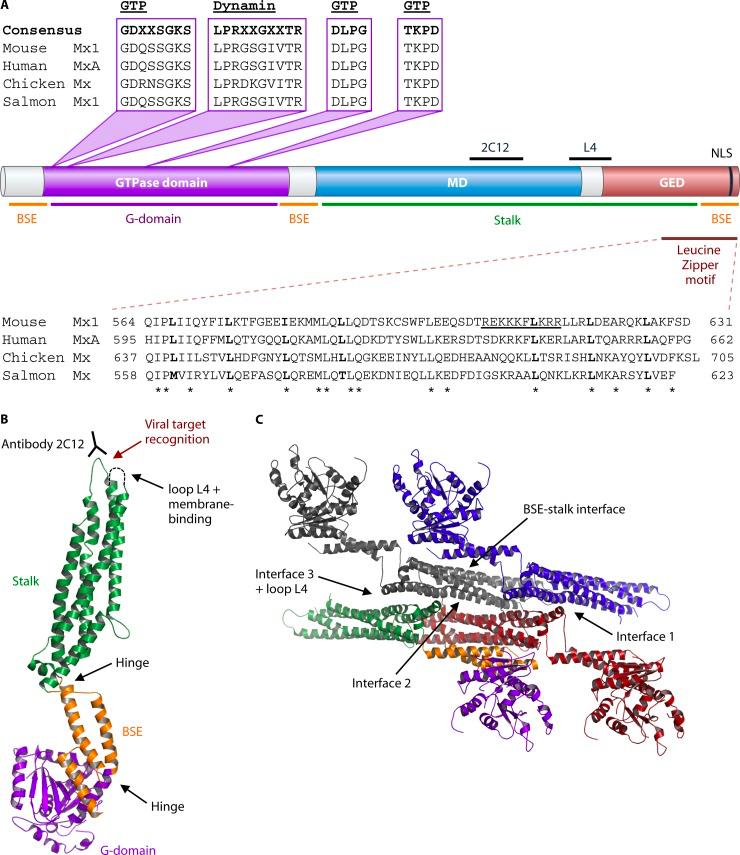
How do Mx proteins perform their antiviral functions?
An intact GTPase domain is required for activity against RNA viruses, but it is not clear if GTP binding is sufficient or GTPase activity is also needed. As the crystal structure of MxA is based on nucleotide-free MxA, it will be interesting to determine the conformational changes caused by GTP binding and GTP hydrolysis in an enzymatically active MxA protein.
The information obtained from the crystal structure of MxA, together with mutational studies, led to the proposition of an antiviral mechanism that depends on the multimeric assembly of Mx proteins. In this model, Mx forms tetramers that further oligomerize into large Mx rings at higher Mx concentrations.
These rings contain GTPase domains on the outside of the ring and stalks pointing inwards. This assembly and oligomerization allow the Mx stalk to interact with viral target structures, e.g., vRNPs, possibly leading to multiple Mx rings wrapping around these structures. Adjacent Mx rings can interact through their GTPase domains, leading to enhanced GTPase activity. This conformation allows cross talk between the GTPase domains and the stalks of neighboring Mx molecules and causes conformational changes leading to constriction of Mx rings and disruption of functional viral vRNPs by a mechano-chemical cycle. In this way, Mx rings could disrupt the association of nucleoprotein complexes with the viral polymerase, thereby inhibiting virus transcription or replication.
These Mx rings could also act to relocalize viral (ribo)nucleoproteins or (nucleo)capsid proteins to perinuclear complexes in a GTPase dependent way, leading to their sequestration and/or degradation. Although this ring-forming model is very appealing, there is no proof that ring formation of Mx around vRNPs or other viral structures actually occurs.